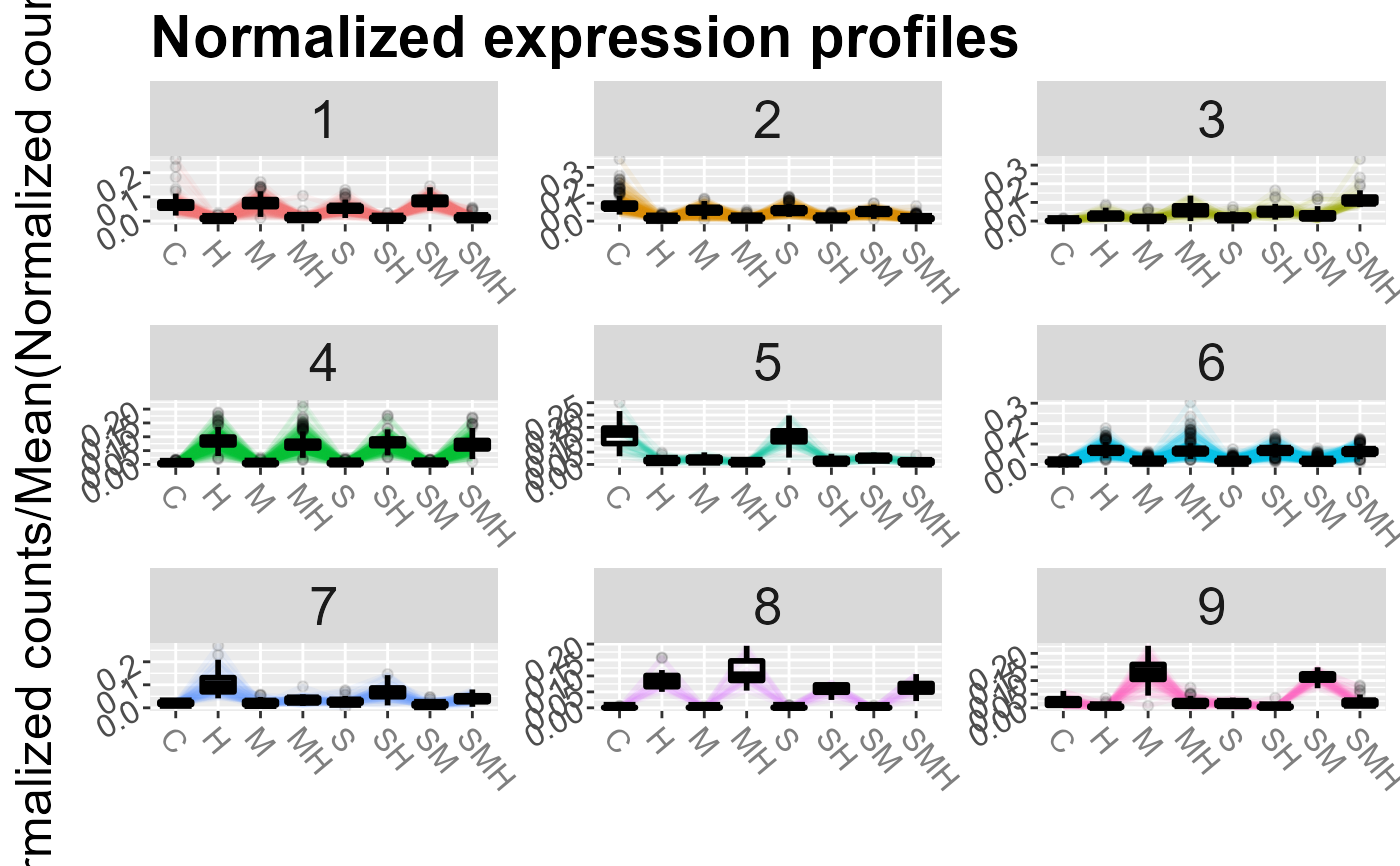
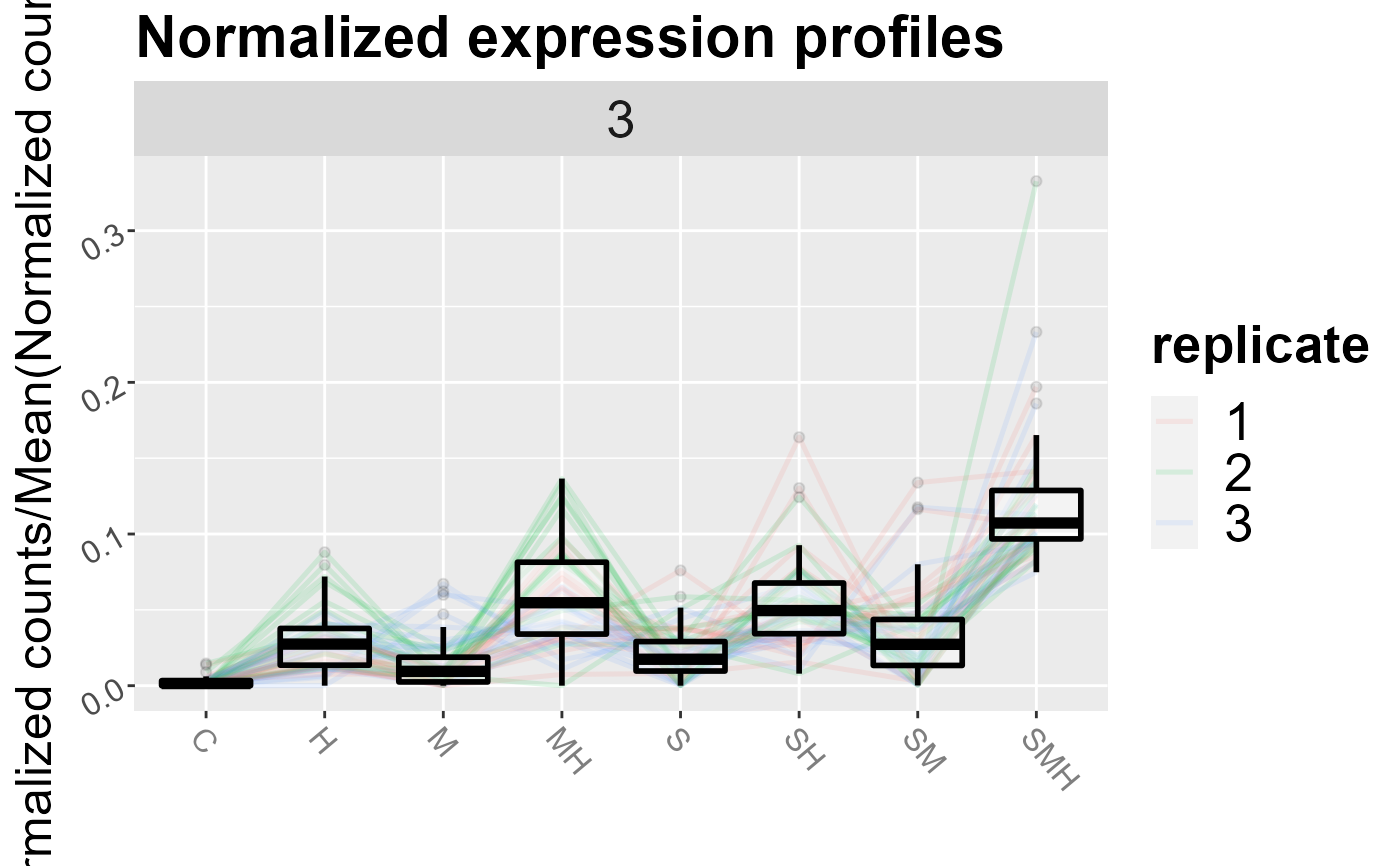
Draw expression profiles of a clustering
draw_profiles( data, membership, conds = unique(stringr::str_split_fixed(colnames(data), "_", 2)[, 1]), expression = "profiles", k = NULL, nrow = 3 )
Arguments
| data | normalized counts with genes as rownames and samples as columns |
|---|---|
| membership | membership item of the coseq object returned by
|
| conds | conditions on which to display clustering profiles. Must be a unique vector containing the conditions you want to consider for gene clustering, without the replicate information (string before the underscore in sample names). Default is all the conditions. |
| expression | if it is set to "profiles" (default), plots expression/sum(expression). if "counts", plots log(Counts+1) |
| k | if NULL (default), plots all the clusters. Else, plot the clusters in the vetcor k. |
| nrow | on how many rows display the cluster profiles if k is NULL |
Examples
data("abiotic_stresses") DIANE::draw_profiles(data = abiotic_stresses$normalized_counts, membership = abiotic_stresses$heat_DEGs_coseq_membership, conds = unique(abiotic_stresses$conditions))DIANE::draw_profiles(data = abiotic_stresses$normalized_counts, membership = abiotic_stresses$heat_DEGs_coseq_membership, conds = unique(abiotic_stresses$conditions), k = 3)